Abstract
The rapid growth of neural network models shared on the internet has made model weights an important data modality. However, this information is underutilized as the weights are uninterpretable, and publicly available models are disorganized. Inspired by Darwin's tree of life, we define the Model Tree which describes the origin of models i.e., the parent model that was used to fine-tune the target model. Similarly to the natural world, the tree structure is unknown. In this paper, we introduce the task of Model Tree Heritage Recovery (MoTHer Recovery) for discovering Model Trees in the ever-growing universe of neural networks. Our hypothesis is that model weights encode this information, the challenge is to decode the underlying tree structure given the weights. Beyond the immediate application of model authorship attribution, MoTHer recovery holds exciting long-term applications akin to indexing the internet by search engines. Practically, for each pair of models, this task requires: i) determining if they are related, and ii) establishing the direction of the relationship. We find that certain distributional properties of the weights evolve monotonically during training, which enables us to classify the relationship between two given models. MoTHer recovery reconstructs entire model hierarchies, represented by a directed tree, where a parent model gives rise to multiple child models through additional training. Our approach successfully reconstructs complex Model Trees, as well as the structure of "in-the-wild" model families such as Llama 2 and Stable Diffusion.
Model Graphs
The number and diversity of neural models shared online have been growing at an unprecedented rate, establishing model weights as an important data modality. On Hugging Face alone there are over 600,000 models, with thousands more added daily. Many of these models are related through a common ancestor (i.e., foundation model) from which they were fine-tuned. We hypothesize that the model weights may encode this relationship.
Motivated by Darwin's tree of life, which describes the relationships between organisms, we analogously define the Model Tree and Model Graph to describe the hereditary relations between models.
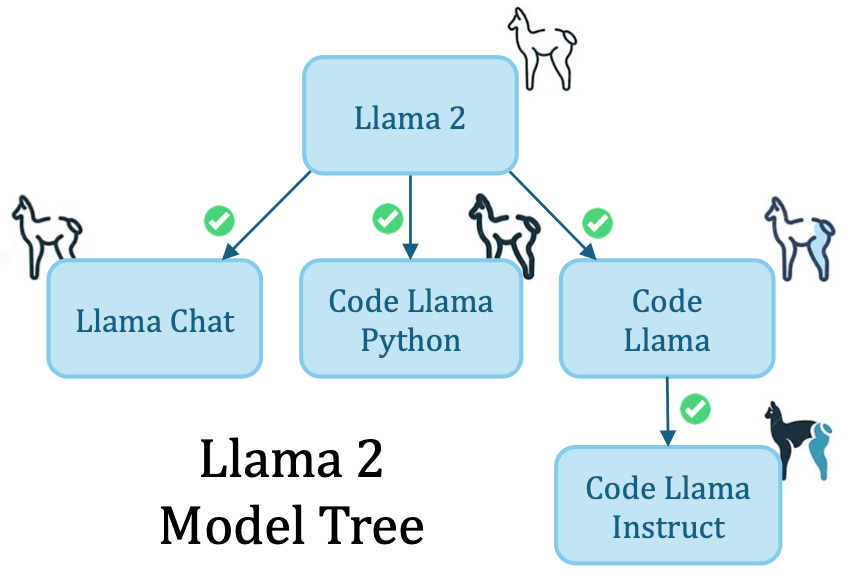 Llama2 Model Tree: We perform MoTHer Recovery in-the-wild, successfully recovering the Llama2 Model Tree with perfect accuracy.
Llama2 Model Tree: We perform MoTHer Recovery in-the-wild, successfully recovering the Llama2 Model Tree with perfect accuracy.
Model Graph Priors
Recovering the structure of a Model Tree requires determining: i) Which nodes are connected and ii) The direction of the connection. We uncover the following properties of model weights:
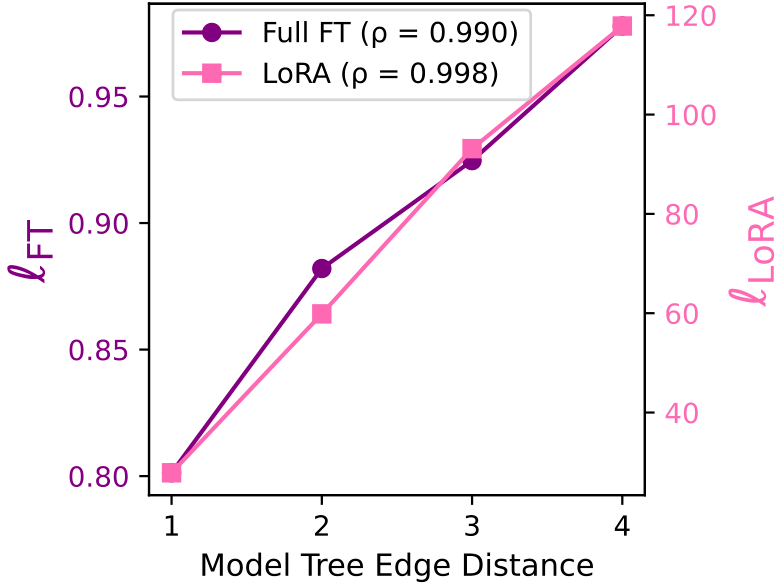 Weight Distance vs. Model Tree Edge Distance: Our weight distances \(\ell_{FT}\) and \(\ell_{LoRA}\) almost perfectly correlate with the number of edges between models in a Model Tree. Thus is a good indicator for determining parent-child relation, i.e., models that were fine-tuned from one another.
Weight Distance vs. Model Tree Edge Distance: Our weight distances \(\ell_{FT}\) and \(\ell_{LoRA}\) almost perfectly correlate with the number of edges between models in a Model Tree. Thus is a good indicator for determining parent-child relation, i.e., models that were fine-tuned from one another.
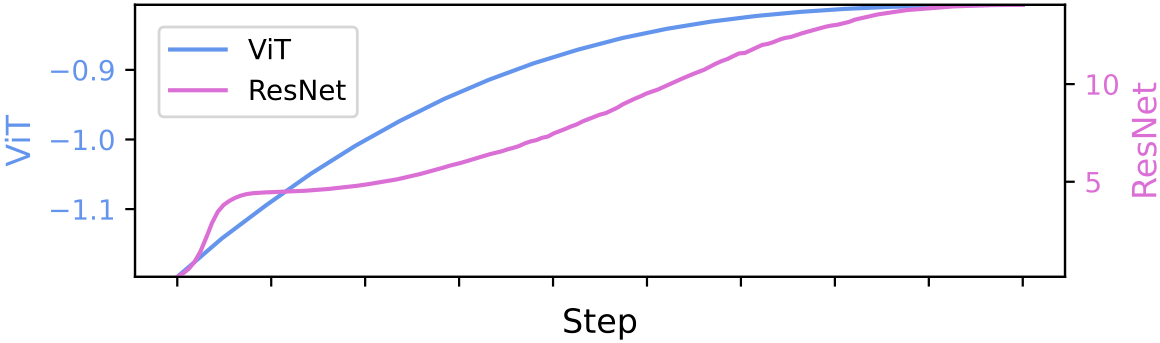
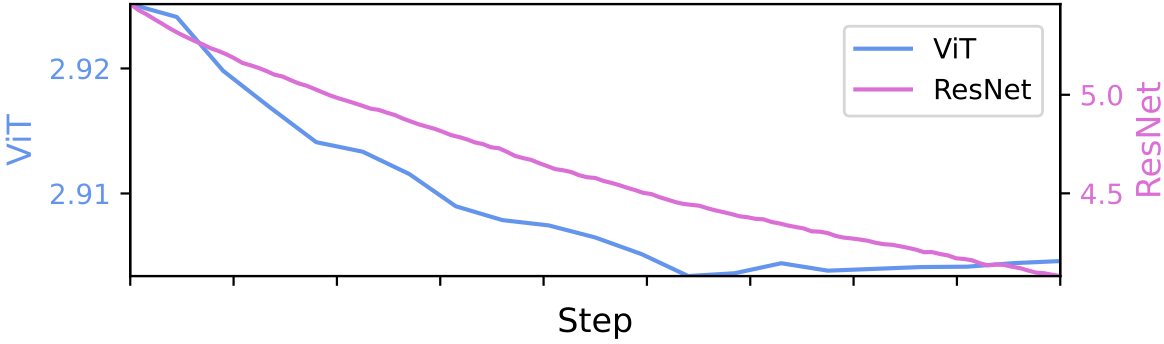 Directional Weight Score: We observe that the number of weight outliers changes monotonically over the course of training. Specifically, we distinguish between a generalization stage (often called pre-training) and a specialization stage (often referred to as fine-tuning). We find that during generalization, the number of weight outliers grows, while during specialization, it diminishes.
Directional Weight Score: We observe that the number of weight outliers changes monotonically over the course of training. Specifically, we distinguish between a generalization stage (often called pre-training) and a specialization stage (often referred to as fine-tuning). We find that during generalization, the number of weight outliers grows, while during specialization, it diminishes.
A Simplified Model Graph
Using these findings, we show a simple algorithm for Model Trees with 3 nodes. We first place the two minimal distance edges. We then take the maximal kurtosis node as the root. If we first cluster the nodes, we can recover all the possible 3 nodes Model Graphs.
MoTHer Recovery: Scaling up Model Graph Recovery
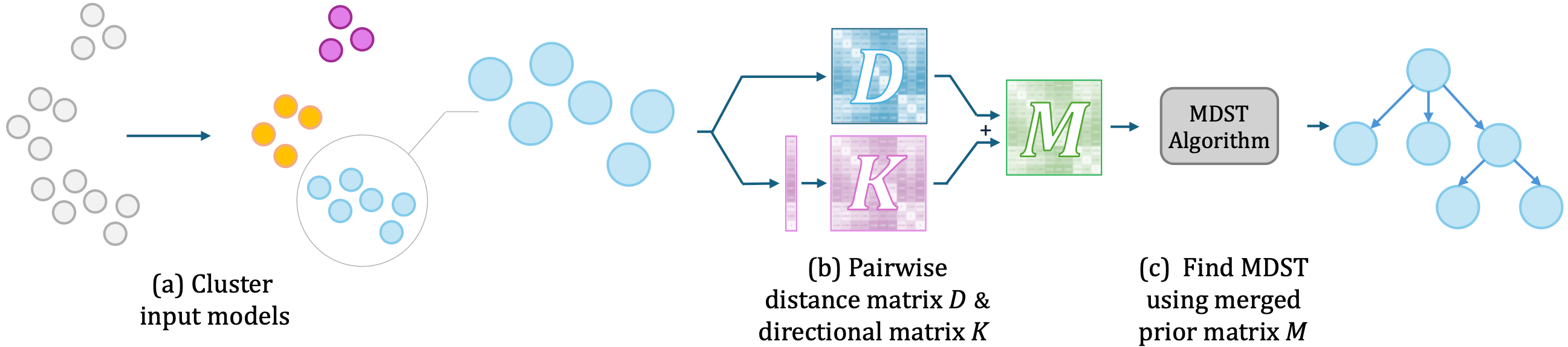
Our algorithm works as follows: (a) Cluster into different Model Trees based on the pairwise weight distances. (b) For each cluster, i) use \(\ell_{FT}\) or \(\ell_{LoRA}\) to create a pairwise distance matrix \(D\) for placing edges, and ii) create a binary directional matrix \(K\) based on the kurtosis to determine the edge direction. (c) To recover the final Model Tree, run a minimum directed spanning tree (MDST) algorithm on the merged prior matrix \(M\). The final recovered Model Graph is the union of the recovered Model Trees.
Results
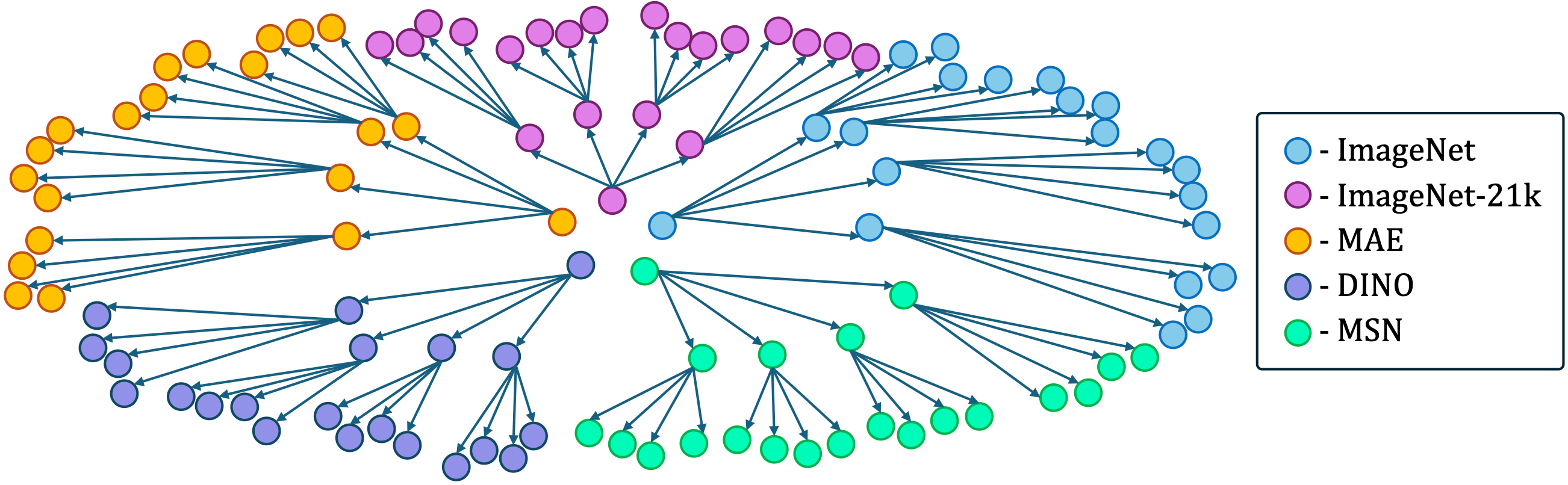
Our method recovered the structure of a ViT-based Model Graph comprising 105 nodes with an 89% accuracy. We visualize the structure of the Model Graph below. Each node in the represents a model that was fine-tuned on different datasets with different configurations. Moreover, when tested on Llama2 our algorithm returned the correct Model Tree.
BibTeX
@inproceedings{
horwitz2025unsupervised,
title={Unsupervised Model Tree Heritage Recovery},
author={Eliahu Horwitz and Asaf Shul and Yedid Hoshen},
booktitle={The Thirteenth International Conference on Learning Representations},
year={2025},
url={https://openreview.net/forum?id=QVj3kUvdvl}
}